Every time cells divide, the preservation, duplication and transmission of their genetic information must be accurately accomplished. In our lab, we study how cells coordinate the repair of DNA damage with major molecular events like DNA replication and chromosome segregation. With a particular focus on homologous recombination, we currently employ a multi-pronged approach that includes biochemistry, yeast genetics and cell biology to focus on the following questions: i) how the biochemical properties of Holliday junction resolvases can bias repair outcome (crossovers vs non-crossovers); ii) how post-translational modifications regulate the biological functions of DNA repair enzymes and iii) how the misregulation of DNA repair activities may interfere with other cellular processes.
Selected Results
|
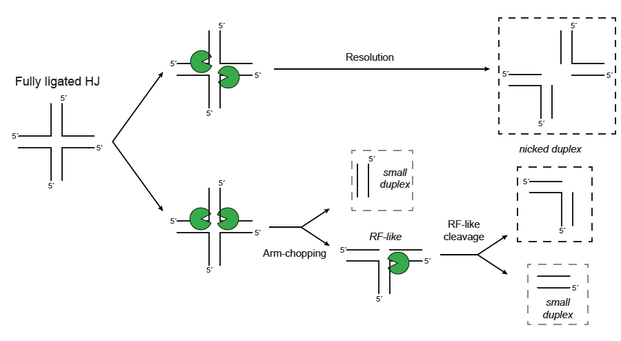
Our in vitro studies of the biochemical properties of the S. cerevisiae Holliday junction resolvase Yen1 has revealed that it shares a number of features with its orthologs in other species, including human GEN1, like its canonical mechanism for the resolution of Holliday junction structures (upper row). However, Yen1 is also endowed with other activities, like an alternative mode of HJ processing termed "arm-chopping" that implies the sequential removal of two helical arms to yield a full-length nicked duplex and two smaller DNA duplexes (lower row). From Carreira et al., 2021. Nucleic Acids Research. |
|
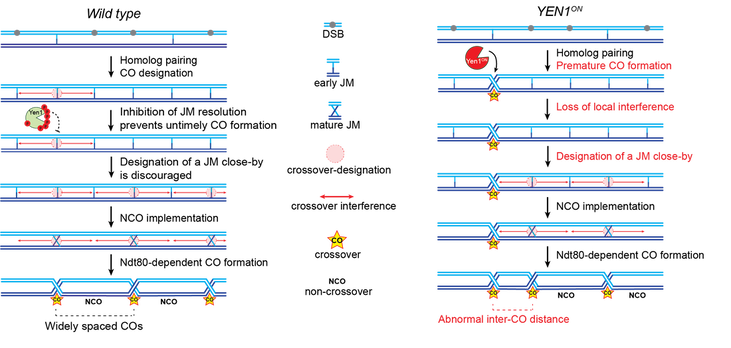
In this collaboration with the Matos lab, we showed that the timely restriction and activation of the Yen1 resolvase contributes not only to the complete removal of persistent joint molecules, but also to the correct spatial patterning of crossovers at a genome-wide level (left panel). This patterning, known as crossover interference is disturbed when Yen1 is prematurely activated, allowing it to processes early joint molecules (right panel). Modified from Arter et al., 2018. Developmental Cell. |
|
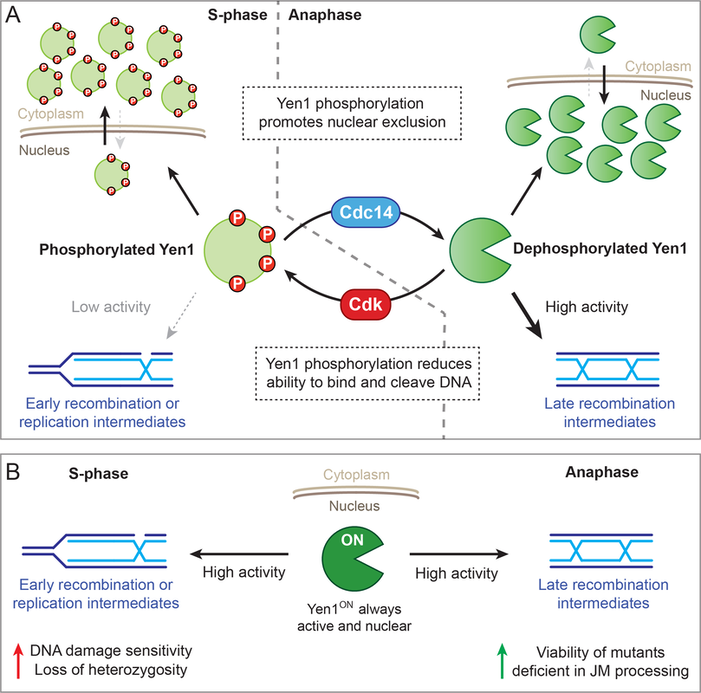
(A) Cdk and Cdc14 control the phosphorylation status of Yen1. In S phase, Cdk phosphorylates Yen1 to (1) promote its nuclear exclusion by impairing its NLS function and, potentially, enhance Msn5-dependent export, and (2) reduce its DNA binding affinity, avoiding the cleavage of S phase-specific DNA structures such as replication or early recombination intermediates. At anaphase, Cdc14 dephosphorylates Yen1 to reinstate a fully functional NLS and potentially block Msn5-mediated export, leading to its nuclear accumulation. Additionally, the removal of phosphate groups increases the DNA binding affinity and catalytic activity of Yen1, allowing it to resolve HR intermediates that have persisted until anaphase. Red circles (P) depict phosphate groups. (B) Yen1ON cannot be turned off or shuttled to the cytoplasm by Cdk. This may lead to the unscheduled and detrimental cleavage of replication or early recombination intermediates. In mutants that accumulate HR intermediates, the constitutive activation of Yen1ON provides an alternative way to process these potentially toxic DNA structures. From Blanco et al., 2014. Molecular Cell |
Groups and networks
- Molecular mechanisms of disease (MeMoEn)
- National network on Chromosome Dynamics and Stability, BFU2015-69142-REDT (CHROMOdyst)
- Health Research Institute of Santiago de Compostela (IDIS)
Collaborators
- Dr. Joao Matos, Institute of Biochemistry, ETH Zurich, Switzerland.
- Dr. Boris Pfander, Max-Planck-Institut für Biochemie, Martisried, Germany.
- Dr. Lumír Krejčí, Masaryk University, Brno, Czech Republic.
- Dra. Mónica Segurado, Instituto de Biología Fundamental y Genómica, Salamanca, Spain.
- Dr. Fernando Moreno Herrero, Centro Nacional de Biotecnología, Madrid, Spain.
- Dr. Rodrigo Bermejo, Centro de Investigaciones Biológicas, Madrid, Spain.
- Dr. Jose Luis García-Pérez, The University of Edinburgh, Edinburgh, Scotland.
- Dra. María de la Fuente, IDIS, Santiago de Compostela.
- Dr. Antonio González, IDIS, Santiago de Compostela.
- Dra. Carmen Rivas, CiMUS, USC and CNB-CSIC.
- Dr. José Manuel Castro Tubío, CiMUS, USC.
- Dr. Fernando Domínguez Puente, CiMUS, USC
- Dr. Miguel Fidalgo, CiMUS, USC
- Dr. Martín Fañanás Mastral, CiQUS, USC
- Dr. Juan Granja Guillán, CiQUS, USC